Tutorial
This is a quick tutorial to get you started with GW and demo some of the main features. This will be a quide for MacOS, but Linux or Windows is more or less the same.
Test data
Firstly, here are some links for some test data:
Launch GW
Once you open the GW app, you will see a terminal window popup:
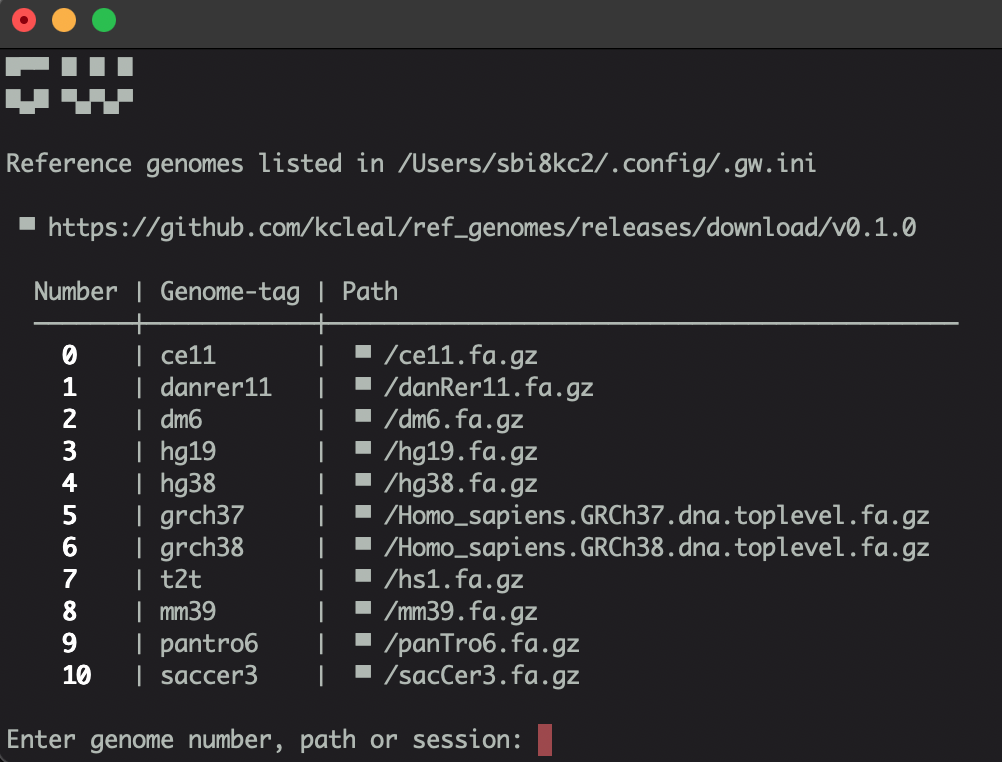
Load the hg19 reference genome by either entering the number, or typing hg19. GW will load an online reference genome, but for much faster performance you can configure GW to use a local file.
Access the Settings and Command box using the popup menus at the left-side of the screen: 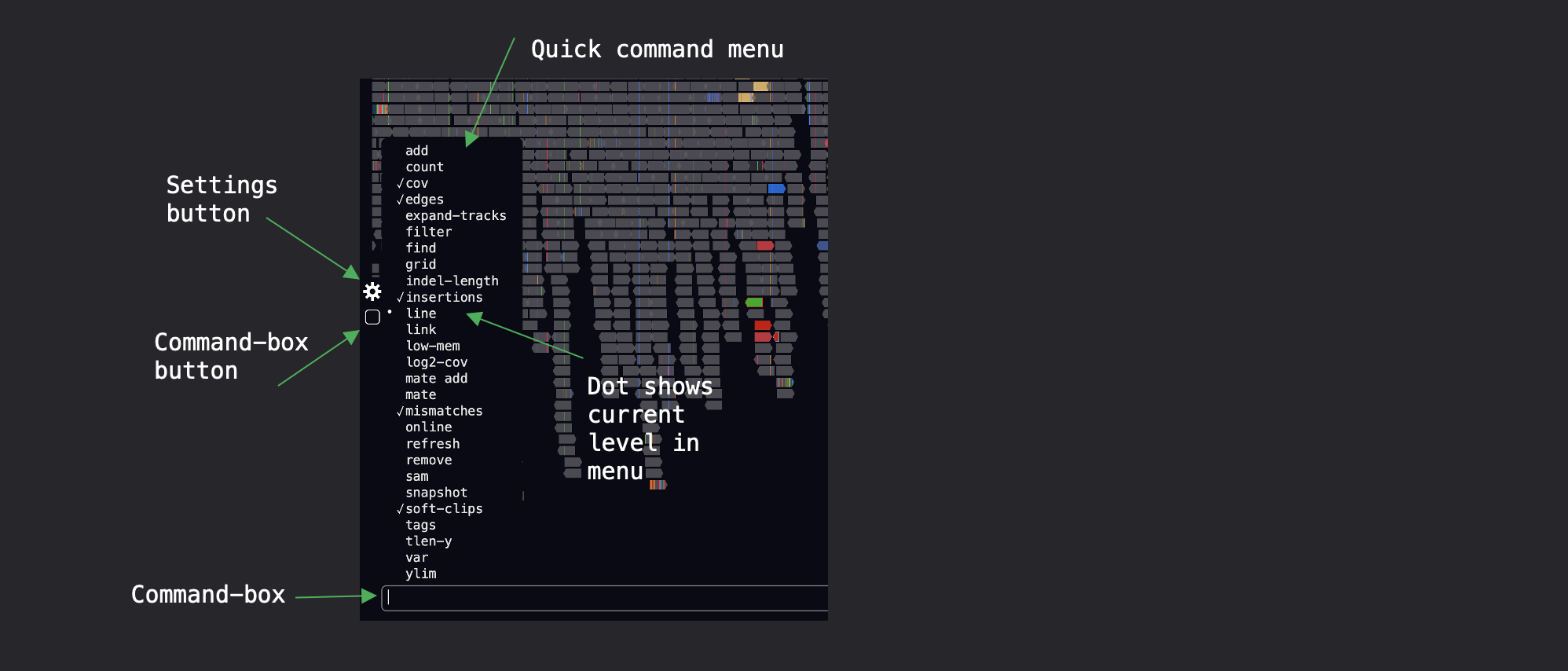
To add a local reference genome, click on the Settings menu (see the cog in the image). Next goto Genomes, and click on the + sign at the top left. You can then enter a genome-tag and the path to your local file.
Loading bams and tracks
Next load some data using one of these options:
- Drag and drop the bam file above (download the index file also!)
-
Use the load command - open the command box and type:
load https://github.com/kcleal/gw/releases/download/v1.0.0/demo1.bam
Navigate to a region by typing this, into the command box. You should now see some data!
chr8:37047270-37055161
Next, load the test bed file (drag-and-drop or use load command):
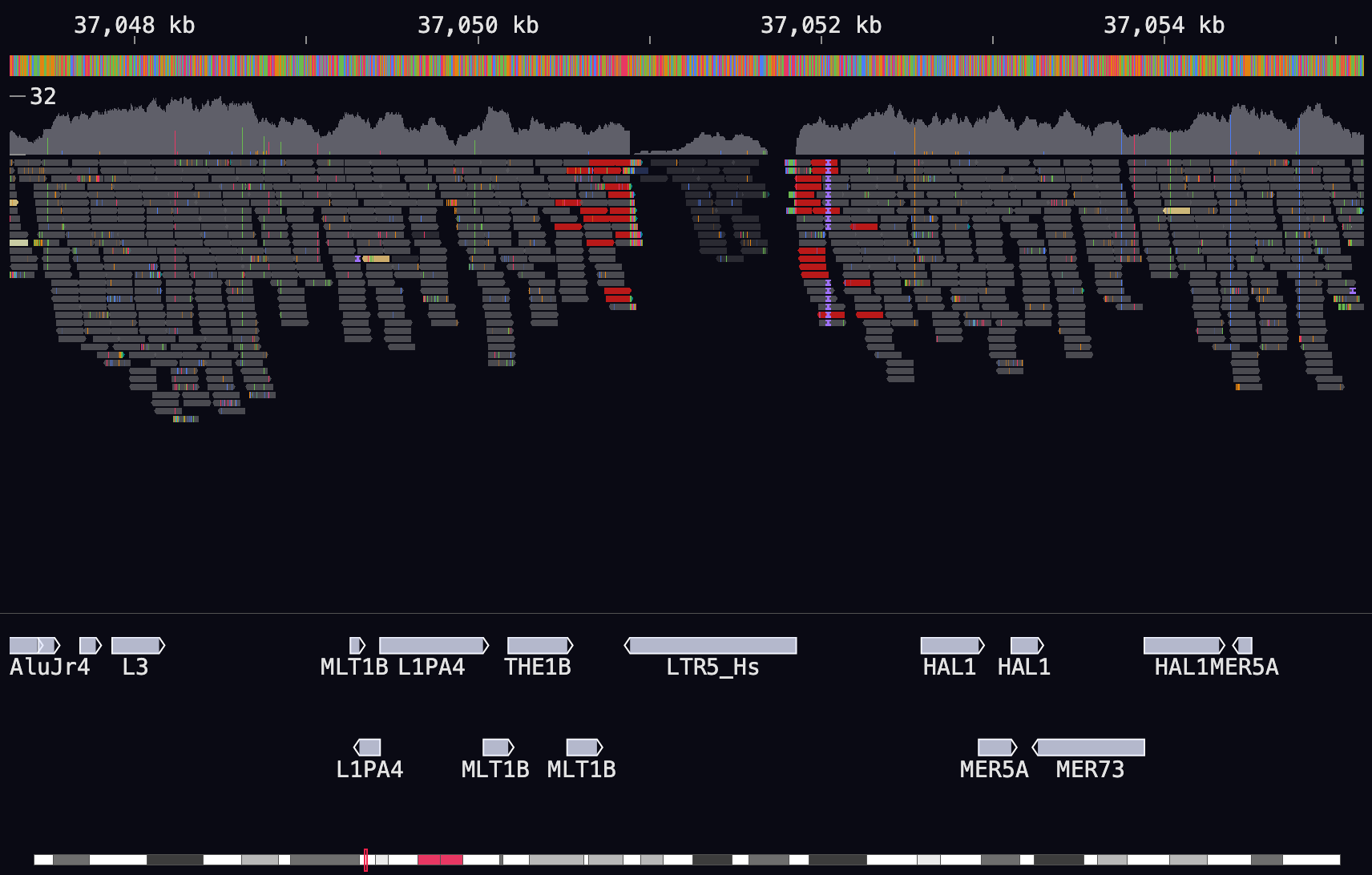
This is the main “alignment-view”. Most screen elements are interactive and display data to the terminal. Navigate around the screen by dragging with the mouse, or using arrow keys.
There are lots of commands available to interact with data. Type help into the command box for a list of whats available.
For now, have a go at using the filter and count commands. In the command box type:
filter pattern = deletion
And then:
count
This will display a count of the deletion-pattern reads to the terminal. To remove the filter, use the refresh or r command.
Loading tiled images / curating variants
GW can also make tiled images of variants from VCF files (or bed files). By default, GW will open VCF/BCF files using the “image-view” mode, creating tiled images from each variant.
Drag-and-drop the test VCF file into the window:
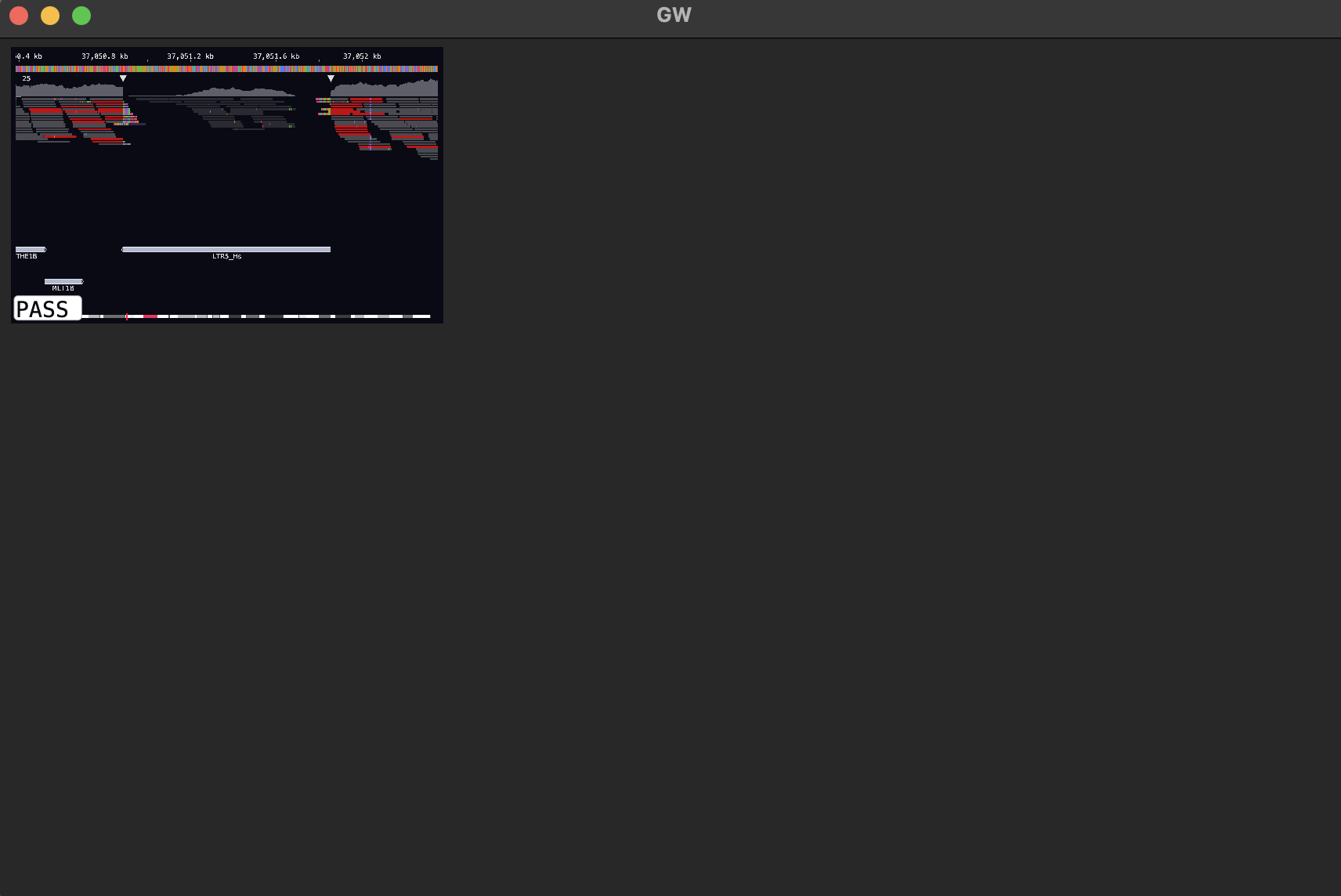
There was only one record in the file, so you only end up with one image!
Clicking on the image will cycle the label. Also you can right-click the image and it will take you to the alignment view where you can explore the data. To go back, just right-click again.
Annotations can be saved and loaded from files. For this, use the save and load commands.
`save labels my_labels.tsv`
This will save the labels to a file. To open them again use:
`load labels my_labels.tsv`