Alignment data
The easiest way to load an alignment file (bam or cram format) is to start GW and drag-and-drop a file into the window. This will open the file and navigate to the first chromosome in the bam file header, showing the first 20 kb.
You can also use the load command from inside GW. Typing load in the command box followed by the path to your bam file (not you can use tab-completion when using this command):
load ~/Desktop/your.bam
Navigate to a new region by typing e.g. chr2 into the command box, or add a new region using add chr2
Using the CLI, bam files are loaded using:
gw hg38 -b your.bam -r chr1:1-20000
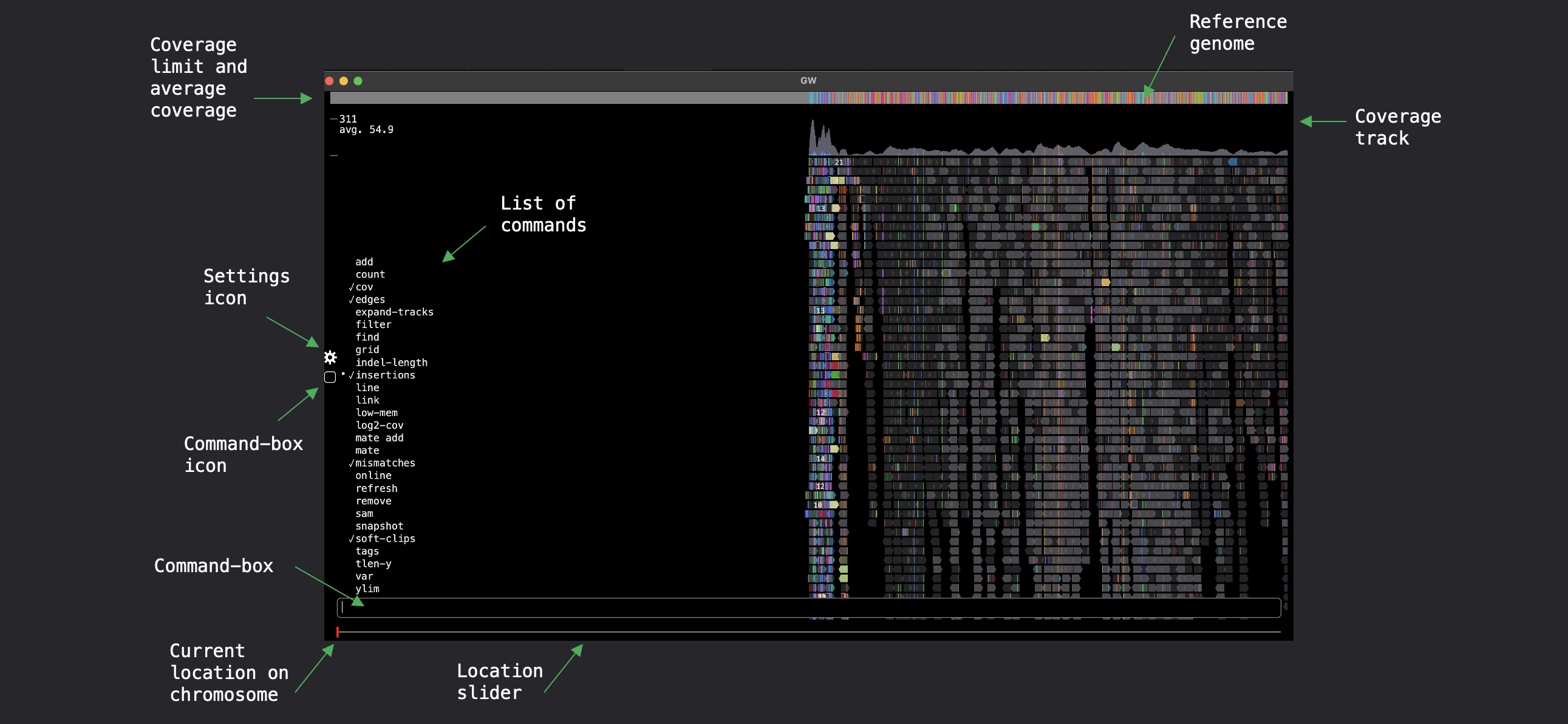
The hg38 argument is a genome-tag listed in the config file and will load a remote reference genome.
For much faster performance, either replace the argument with the path to a local file, or change the config file to point to a local file rather than a remote reference genome (see Genomes section for more detail). This command will open a GW window that can be used interactively with the mouse and keyboard. Note multiple -b and -r options can be used.
Genome tags
If you have a genome-tag configured in your config file it is possible to provide the alignment file as a single argument:
gw your.bam
GW will read the bam file header and infer the correct reference genome to load by comparing with the references listed in your config file. If you are using the desktop application this feature allows you to use the Open with function (found by right-clicking on a bam file).