Commands
To open the command box press the forward slash or click on the button found on the menu at the left-hand-side of the screen:
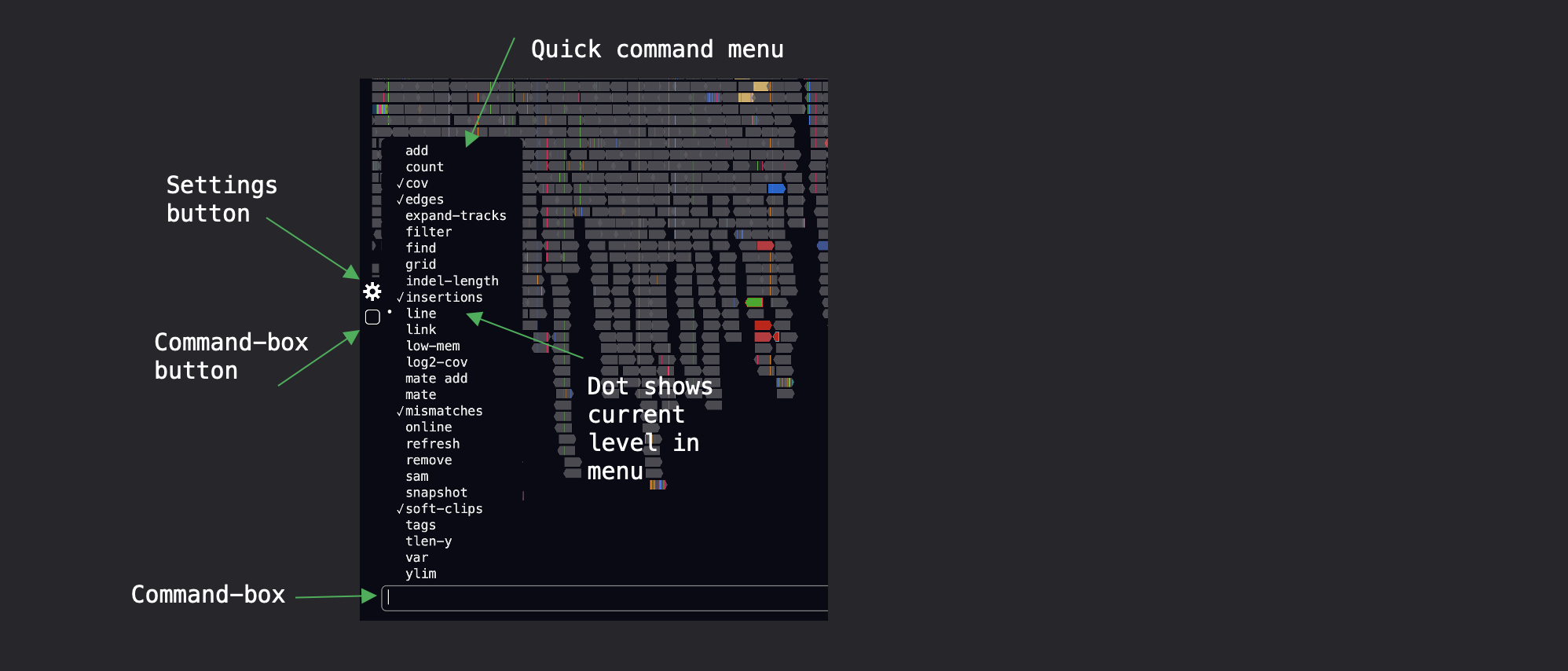
Type help to print a quick help summary to the terminal, or for more information about each command type man [COMMAND] into the box, where COMMAND is the name of the command e.g.
man ylim
If you type some text that is not recognised as a command, then the text will be output in the terminal. This can be useful for note-taking.
Pressing the UP ARROW key will cycle through your command history.
The list of available commands are given below.
Table of contents
- [locus / feature]
- add
- colour
- count
- cov
- edges
- expand-tracks
- filter
- find, f
- goto
- grid
- header
- indel-length
- insertions, ins
- line
- link, l
- log2-cov
- mate
- mismatches, mm
- mods
- online
- quit, q
- refresh, r
- remove, rm
- roi
- sam
- save
- settings
- snapshot, s
- soft-clips, sc
- sort
- tags
- theme
- tlen-y
- var, v
- ylim
Navigate to a genomic locus, or feature of interest. You can use chromosome names or chromosome coordinates to change the current view point to a new location.
chr1:1-20000
chr1
chr1:10000
Additionally, if you have tracks loaded, such as a bed file of genes, you can type the name of a gene or feature into the command box to navigate to that feature. Note, this feature only really works with non-indexed files, because when using indexed files, GW will only load the data that is on-screen, rather than the whole file.
hTERT
This will add a new region view to the right-hand-side of your screen.
add chr1:1-20000
Set the (alpha, red, green, blue) colour for one of the plot elements. Elements are selected by name (see below) and values are in the range [0, 255]. For example colour fcNormal 255 255 0 0 sets face-colour of normal reads to red.
bgPaint - background paint
bgMenu - background of menu
fcNormal - face-colour normal reads
fcDel - face-colour deletion pattern reads
fcDup - face-colour duplication pattern reads
fcInvF - face-colour inversion-forward pattern reads
fcInvR - face-colour inversion-reverse pattern reads
fcTra - face-colour translocation pattern reads
fcIns - face-colour insertion blocks
fcSoftClips - face-colour soft-clips when zoomed-out
fcA - face-colour A mismatch
fcT - face-colour T mismatch
fcC - face-colour C mismatch
fcG - face-colour G mismatch
fcN - face-colour N mismatch
fcCoverage - face-colour coverage track
fcTrack - face-colour tracks
fcBigWig - face-colour bigWig files
fcNormal0 - face-colour normal reads with mapq=0
fcDel0 - face-colour deletion pattern reads with mapq=0
fcDup0 - face-colour duplication pattern reads with mapq=0
fcInvF0 - face-colour inversion-forward pattern reads with mapq=0
fcInvR0 - face-colour inversion-reverse pattern reads with mapq=0
fcTra0 - face-colour translocation pattern reads with mapq=0
fcIns0 - face-colour insertion blocks with mapq=0
fcSoftClips0 - face-colour soft-clips when zoomed-out with mapq=0
fcMarkers - face-colour of markers
fc5mc - face-colour of 5-Methylcytosine
fc5hmc - face-colour of 5-Hydroxymethylcytosine
ecMateUnmapped - edge-colour mate unmapped reads
ecSplit - edge-colour split reads
ecSelected - edge-colour selected reads
lcJoins - line-colour of joins
lcCoverage - line-colour of coverage profile
lcLightJoins - line-colour of lighter joins
lcLabel - line-colour of labels
lcBright - line-colour of bright edges
tcDel - text-colour of deletions
tcIns - text-colour of insertions
tcLabels - text-colour of labels
tcBackground - text-colour of background
Counts the visible reads in each view. A summary output will be displayed for each view on the screen. Optionally a filter expression may be added to the command. See the section for ‘filter’ for mote details.
count
...
File HG002.bwa.bam
Region chr1:1-20000
Total 4220
Paired 4220
Proper-pair 3791
Read-unmapped 0
Mate-unmapped 6
Read-reverse 2024
Mate-reverse 2107
First-in-pair 2118
Second-in-pair 2102
Not-primary 0
Fails-qc 0
Duplicate 60
Supplementary 76
Deletion-pattern 6
Duplication-pattern 3
Translocation-pattern 356
F-inversion-pattern 26
R-inversion-pattern 40
Count after applying a filter expression:
count flag & 2
count flag & proper-pair
Toggles the coverage track, turning it off/on.
Toggles the edges of tracks, turning them off/on. Edge-highlights are used to indicate split-reads, or reads that have an unmapped mate-pair.
Toggle expand-tracks. Features in the tracks panel are expanded so overlapping features can be seen.
filter
Filter visible reads.
Reads can be filtered using an expression ‘{property} {operation} {value}’ (the white-spaces are also needed). For example, here are some useful expressions:
filter mapq >= 20
filter flag & 2048
filter seq contains TTAGGG
Properties are interpreted as either Numeric-like or String-like.
Numeric Properties
These properties can be used with numeric operators:
mapq- Mapping qualityflag- Bit flag~flag- Bit-wise NOT flagtlen- Template-lengthabs-tlen- Absolute template-lengthpos- Alignment start positionref-end- Alignment end positionpnext- Position of mateseq-len- Sequence lengthNM,CM,FI,HO,MQ,SM,TC,UQ,AS,HP- BAM tags
String Properties
These properties can be used with string operators:
name- Read namername- Chromosome namernext- Chromosome name of matecigar- CIGAR string of alignmentseq- Sequence of this alignmentseq-rc- Reverse-complement sequence of this alignmentRG,BC,BX,RX,LB,MD,MI,PU,SA,MC- BAM tags
Numeric Operators
Numeric types can be combined with these operators:
==- Equal to; or use=oreq!=- Not equal to; orne>- Greater than;gt<- Less than;lt>=- Greater-or-equal;ge<=- Less-or-equal;le
String Operators
String types can be combined with these operators:
==- Equal to; or use=oreq!=- Not equal to; ornecontains- String contains substringomit- Removes read if string contains substring
Missing Values
If you want to filter using missing values, you can use none or '', e.g:
filter SA == none # Only reads with no SA tag are kept
Pattern Filtering
Reads can be filtered on their mapping orientation/pattern:
filter pattern == del # deletion-like pattern
filter pattern == inv_f # inversion-forward
filter pattern == inv_r # inversion-reverse
filter pattern != tra # translocation
Bitwise Flag Names
Bitwise flags can also be applied with named values:
paired, proper-pair, unmapped, munmap, reverse, mreverse, read1, read2, secondary, dup, supplementary
Examples:
filter paired
filter read1
Chaining Expressions
Expressions can be chained together providing all expressions are ‘AND’ or ‘OR’ blocks:
filter mapq >= 20 and mapq < 30
filter mapq >= 20 or flag & supplementary
Panel-Specific Filtering
You can apply filters to specific panels using array indexing notation:
filter mapq > 0 [:, 0] # All rows, column 0 (all bams, first region only)
filter mapq > 0 [0, :] # Row 0, all columns (the first bam only, all regions)
filter mapq > 0 [1, -1] # Row 1, last column
Example Commands
Here are some example filtering commands:
filter mapq >= 20 # only reads with mapping quality >= 20 will be shown
filter flag & 2048 # only supplementary alignments are shown
filter supplementary # same as above
filter ~supplementary # supplementary reads will be removed
filter seq contains TTAGGG # Only reads with TTAGGG kmer will be shown
filter seq omit AAAAAA # Reads with this kmer will be removed
filter mapq > 30 and ~dup # also removes duplicate reads
filter mapq > 10 or seq-len > 100 # multiple conditions with OR
Find a read with target name.
All alignments with the same name will be highlighted with a bright edge border.
find D00360:18:H8VC6ADXX:1:1107:5538:24033
Navigate to a locus or track feature.
This moves the current region to a new view point, or adds a new view point if one is not already open. You can also specify a feature name from one of the loaded tracks. Searching for track features works mainly for non-indexed files that are loaded into memory during startup.
goto chr1
goto hTERT
Set the grid size for viewing image tiles.
grid 8x8 # this will display 64 image tiles
Prints the header of the current selected bam to the terminal. Use header names to print only the @SQ lines.
header # print the full header
header names # prints only the @SQ lines (chromosome names)
Set the minimum indel-length.
Indels (gaps in alignments) will be labelled with text if they have length ≥ ‘indel-length
indel-length 30
Toggle insertions. Insertions smaller than ‘indel-length’ are turned on or off.
Toggles a vertical line drawn over cursor position.
Link alignments.
This will change how alignments are linked, options are ‘none’, ‘sv’, ‘all’. Linking by ‘sv’ will draw links between alignments that have either a discordant flag, or have a supplementary mapping.
Toggle log2-coverage. The coverage track will be scaled by log2.
Goto mate alignment.
Either moves the current view to the mate locus, or adds a new view of the mate locus.
mate
mate add
Toggle mismatches. Mismatches with the reference genome are turned on or off.
Toggle base modifications. Mods are turned on or off.
Show links to online browsers for the current region.
This function is mainly implemented for human reference genomes including hg19/GRCh37, hg38/GRCh38 and hs1/t2t. If you are not using a genome tag, you can add one as an additional argument:
online
online GRCh37
For human references, links to databases with the same region as the current view are provided. Supported databases include:
| UCSC Genome Browser link |
| DECIPHER Genome Browser link |
| Gnomad Browser link |
| Ensemble Genome Browser link |
| NCBI Genome Data Viewer link |
If you would like to see more databases added, please get in touch.
Simply quits GW
Refresh the screen and re-draws everything. All filters will also be removed.
Remove a region, bam or track by index.
To remove a bam or track add a ‘bam’ or ‘track’ prefix.
rm 0 # this will remove REGION at index 0
rm bam1 # this will remove BAM FILE at index 1
rm track2 # removes track at index 2
Add a region of interest. If no arguments are added, the visible region will be added to the roi track.
roi
roi chr1:1-20000
Print the selected read to the terminal, or save in a file.
First select a read using the mouse then type sam. Alternatively, use the the > key to redirect to a file, or >> to append to a file. Note, the header will also be written, and the output file will be unsorted:
sam
sam > single_read.sam # save in sam format
sam >> multiple_reads.bam # Append read to a bam file
sam >> multiple_reads.cram # Append in cram format
This command can be used to save visible reads, a snapshot, or a session file. The suffix is used to determine the output format:
save reads.bam # visible reads output to bam file
save reads.cram # saved in cram format
save reads.bam [:, 1] # Pane-indexing supported, see filter command for more details
save region.png # Image saved, same as snapshot command
save current_session.ini # Save a session file
Opens the settings panel.
Save an image of the screen.
If no name is provided, the image name will be ‘chrom_start_end.png’ and will be saved in the current working directory, or if you are in tiled-mode, the image name will be ‘index_start_end.png’
snapshot
snapshot my_image.png
If you have a vcf/bcf open in ‘single’ mode (not ‘tiled’) it is also possible to parse info from the vcf record. Use curley braces to select what fields to use in the filename:
snapshot {pos}_{qual}.png # parse the position and qual fields
snapshot {info.SU}.png # parse SU from info field
s {format[samp1].SU}.png # samp1 sample, SU column from format field
Valid fields are chrom, pos, id, ref, alt, qual, filter, info, format. Just to note, you can press the repeat key (R by default) to repeat the last command, which can save typing this command over and over.
Soft-clipped bases or hard-clips are turned on or off.
Sort reads by strand, haplotype (defined by HP tag in bam file), or pos. Sorting by position also has a hotkey - pressing S will sort based on cursor position.
sort hap
sort strand
sort 6400234 # Sorting based on genomic position
sort strand 120000 # By strand and then position
sort hap 120000 # Haplotype then position
This will print all the tags of the selected read (select a read with the mouse first).
Alternatively supply a list of tags to print out:
tags NM RG
Switch the theme.
Currently, ‘igv’, ‘dark’ or ‘slate’ themes are supported.
theme slate
Toggle tlen-y option.
This will scale the read position by template length, so will apply to paired-end reads only. Reads with a shorter template-length will be drawn towards the top of the screen, and reads with a longer template-length will be drawn below.
You can set a maximum y-limit on the template-length by adding a number argument:
tlen-y
tlen-y 50000
Print variant information.
This function requires a VCF file to be opened as a variant file, which can be added using the -v option on command line, or by dragging-and-dropping a VCF into the window.
Using var will print the full record of the selected variant (use mouse to select). If you are viewing a VCF then columns can be parsed e.g:
var pos # position
var info.SU # SU column from info
v chrom pos info.SU # list of variables to print
v format.SU # SU column from format
v format[samp1].SU # using sample name to select SU
Valid fields are chrom, pos, id, ref, alt, qual, filter, info, format. Just to note, you can press ENTER to repeat the last command, which can save typing this command over and over.
Set the y limit. The y limit is the maximum height of stacked reads shown on the drawing.
ylim 100